2. Comparing
A set of informative, discriminating and independent features is important for
a good classification of record pairs into matching and distinct pairs. The
recordlinkage.Compare class and its methods can be used to compare records
pairs. Several comparison methods are included such as string similarity
measures, numerical measures and distance measures.
recordlinkage.Compare object
- class recordlinkage.Compare(features=[], n_jobs=1, indexing_type='label', **kwargs)
Class to compare record pairs with efficiently.
Class to compare the attributes of candidate record pairs. The
Compareclass has methods likestring,exactandnumericto initialise the comparing of the records. Thecomputemethod is used to start the actual comparing.Example
Consider two historical datasets with census data to link. The datasets are named
census_data_1980andcensus_data_1990. The MultiIndexcandidate_pairscontains the record pairs to compare. The record pairs are compared on the first name, last name, sex, date of birth, address, place, and income:# initialise class comp = recordlinkage.Compare() # initialise similarity measurement algorithms comp.string('first_name', 'name', method='jarowinkler') comp.string('lastname', 'lastname', method='jarowinkler') comp.exact('dateofbirth', 'dob') comp.exact('sex', 'sex') comp.string('address', 'address', method='levenshtein') comp.exact('place', 'place') comp.numeric('income', 'income') # the method .compute() returns the DataFrame with the feature vectors. comp.compute(candidate_pairs, census_data_1980, census_data_1990)
- Parameters:
features (list) – List of compare algorithms.
n_jobs (integer, optional (default=1)) – The number of jobs to run in parallel for comparing of record pairs. If -1, then the number of jobs is set to the number of cores.
indexing_type (string, optional (default='label')) – The indexing type. The MultiIndex is used to index the DataFrame(s). This can be done with pandas
.locor with.iloc. Use the value ‘label’ to make use of.locand ‘position’ to make use of.iloc. The value ‘position’ is only available when the MultiIndex consists of integers. The value ‘position’ is much faster.
- add(model)
Add a compare method.
This method is used to add compare features.
- Parameters:
model (list, class) – A (list of) compare feature(s) from
recordlinkage.compare.
- compute(pairs, x, x_link=None)
Compare the records of each record pair.
Calling this method starts the comparing of records.
- Parameters:
pairs (pandas.MultiIndex) – A pandas MultiIndex with the record pairs to compare. The indices in the MultiIndex are indices of the DataFrame(s) to link.
x (pandas.DataFrame) – The DataFrame to link. If x_link is given, the comparing is a linking problem. If x_link is not given, the problem is one of duplicate detection.
x_link (pandas.DataFrame, optional) – The second DataFrame.
- Returns:
pandas.DataFrame – A pandas DataFrame with feature vectors, i.e. the result of comparing each record pair.
- compare_vectorized(comp_func, labels_left, labels_right, *args, **kwargs)
Compute the similarity between values with a callable.
This method initialises the comparing of values with a custom function/callable. The function/callable should accept numpy.ndarray’s.
Example
>>> comp = recordlinkage.Compare() >>> comp.compare_vectorized(custom_callable, 'first_name', 'name') >>> comp.compare(PAIRS, DATAFRAME1, DATAFRAME2)
- Parameters:
comp_func (function) – A comparison function. This function can be a built-in function or a user defined comparison function. The function should accept numpy.ndarray’s as first two arguments.
labels_left (label, pandas.Series, pandas.DataFrame) – The labels, Series or DataFrame to compare.
labels_right (label, pandas.Series, pandas.DataFrame) – The labels, Series or DataFrame to compare.
*args – Additional arguments to pass to callable comp_func.
**kwargs – Additional keyword arguments to pass to callable comp_func. (keyword ‘label’ is reserved.)
label ((list of) label(s)) – The name of the feature and the name of the column. IMPORTANT: This argument is a keyword argument and can not be part of the arguments of comp_func.
- exact(*args, **kwargs)
Compare attributes of pairs exactly.
Shortcut of
recordlinkage.compare.Exact:from recordlinkage.compare import Exact indexer = recordlinkage.Compare() indexer.add(Exact())
- string(*args, **kwargs)
Compare attributes of pairs with string algorithm.
Shortcut of
recordlinkage.compare.String:from recordlinkage.compare import String indexer = recordlinkage.Compare() indexer.add(String())
- numeric(*args, **kwargs)
Compare attributes of pairs with numeric algorithm.
Shortcut of
recordlinkage.compare.Numeric:from recordlinkage.compare import Numeric indexer = recordlinkage.Compare() indexer.add(Numeric())
- geo(*args, **kwargs)
Compare attributes of pairs with geo algorithm.
Shortcut of
recordlinkage.compare.Geographic:from recordlinkage.compare import Geographic indexer = recordlinkage.Compare() indexer.add(Geographic())
- date(*args, **kwargs)
Compare attributes of pairs with date algorithm.
Shortcut of
recordlinkage.compare.Date:from recordlinkage.compare import Date indexer = recordlinkage.Compare() indexer.add(Date())
Algorithms
- class recordlinkage.compare.Exact(left_on, right_on, agree_value=1, disagree_value=0, missing_value=0, label=None)
Compare the record pairs exactly.
This class is used to compare records in an exact way. The similarity is 1 in case of agreement and 0 otherwise.
- Parameters:
left_on (str or int) – Field name to compare in left DataFrame.
right_on (str or int) – Field name to compare in right DataFrame.
agree_value (float, str, numpy.dtype) – The value when two records are identical. Default 1. If ‘values’ is passed, then the value of the record pair is passed.
disagree_value (float, str, numpy.dtype) – The value when two records are not identical.
missing_value (float, str, numpy.dtype) – The value for a comparison with a missing value. Default 0.
- compute(pairs, x, x_link=None)
Compare the records of each record pair.
Calling this method starts the comparing of records.
- Parameters:
pairs (pandas.MultiIndex) – A pandas MultiIndex with the record pairs to compare. The indices in the MultiIndex are indices of the DataFrame(s) to link.
x (pandas.DataFrame) – The DataFrame to link. If x_link is given, the comparing is a linking problem. If x_link is not given, the problem is one of duplicate detection.
x_link (pandas.DataFrame, optional) – The second DataFrame.
- Returns:
pandas.Series, pandas.DataFrame, numpy.ndarray – The result of comparing record pairs (the features). Can be a tuple with multiple pandas.Series, pandas.DataFrame, numpy.ndarray objects.
- class recordlinkage.compare.String(left_on, right_on, method='levenshtein', threshold=None, missing_value=0.0, label=None)
Compute the (partial) similarity between strings values.
This class is used to compare string values. The implemented algorithms are: ‘jaro’,’jarowinkler’, ‘levenshtein’, ‘damerau_levenshtein’, ‘qgram’ or ‘cosine’. In case of agreement, the similarity is 1 and in case of complete disagreement it is 0. The Python Record Linkage Toolkit uses the jellyfish package for the Jaro, Jaro-Winkler, Levenshtein and Damerau- Levenshtein algorithms.
- Parameters:
left_on (str or int) – The name or position of the column in the left DataFrame.
right_on (str or int) – The name or position of the column in the right DataFrame.
method (str, default 'levenshtein') – An approximate string comparison method. Options are [‘jaro’, ‘jarowinkler’, ‘levenshtein’, ‘damerau_levenshtein’, ‘qgram’, ‘cosine’, ‘smith_waterman’, ‘lcs’]. Default: ‘levenshtein’
threshold (None, float, tuple of floats) – A threshold value. All approximate string comparisons higher or equal than this threshold are 1. Otherwise 0. If None, it returns the float string comparison value instead of 0 or 1. Default None.
missing_value (numpy.dtype) – The value for a comparison with a missing value. Default 0.
label (list, str, int) – The identifying label(s) for the returned values. Default None.
- compute(pairs, x, x_link=None)
Compare the records of each record pair.
Calling this method starts the comparing of records.
- Parameters:
pairs (pandas.MultiIndex) – A pandas MultiIndex with the record pairs to compare. The indices in the MultiIndex are indices of the DataFrame(s) to link.
x (pandas.DataFrame) – The DataFrame to link. If x_link is given, the comparing is a linking problem. If x_link is not given, the problem is one of duplicate detection.
x_link (pandas.DataFrame, optional) – The second DataFrame.
- Returns:
pandas.Series, pandas.DataFrame, numpy.ndarray – The result of comparing record pairs (the features). Can be a tuple with multiple pandas.Series, pandas.DataFrame, numpy.ndarray objects.
- class recordlinkage.compare.Numeric(left_on, right_on, method='linear', offset=0.0, scale=1.0, origin=0.0, missing_value=0.0, label=None)
Compute the (partial) similarity between numeric values.
This class is used to compare numeric values. The implemented algorithms are: ‘step’, ‘linear’, ‘exp’, ‘gauss’ or ‘squared’. In case of agreement, the similarity is 1 and in case of complete disagreement it is 0. The implementation is similar with numeric comparing in ElasticSearch, a full- text search tool. The parameters are explained in the image below (source ElasticSearch, The Definitive Guide)
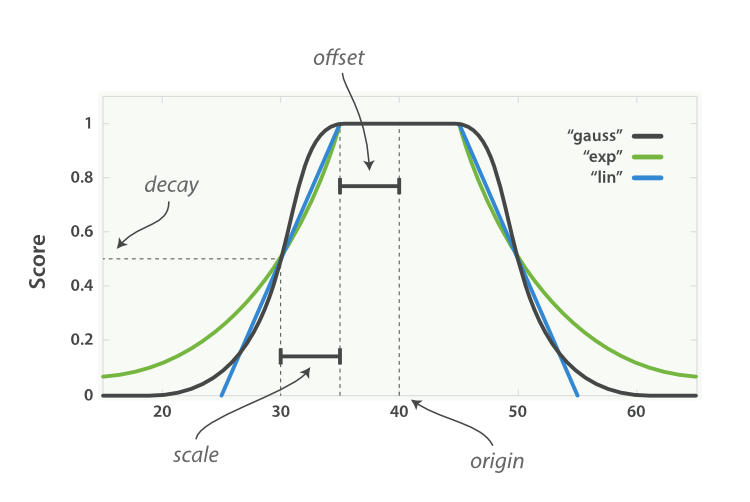
- Parameters:
left_on (str or int) – The name or position of the column in the left DataFrame.
right_on (str or int) – The name or position of the column in the right DataFrame.
method (float) – The metric used. Options ‘step’, ‘linear’, ‘exp’, ‘gauss’ or ‘squared’. Default ‘linear’.
offset (float) – The offset. See image above.
scale (float) – The scale of the numeric comparison method. See the image above. This argument is not available for the ‘step’ algorithm.
origin (float) – The shift of bias between the values. See image above.
missing_value (numpy.dtype) – The value if one or both records have a missing value on the compared field. Default 0.
Note
Numeric comparing can be an efficient way to compare date/time variables. This can be done by comparing the timestamps.
- compute(pairs, x, x_link=None)
Compare the records of each record pair.
Calling this method starts the comparing of records.
- Parameters:
pairs (pandas.MultiIndex) – A pandas MultiIndex with the record pairs to compare. The indices in the MultiIndex are indices of the DataFrame(s) to link.
x (pandas.DataFrame) – The DataFrame to link. If x_link is given, the comparing is a linking problem. If x_link is not given, the problem is one of duplicate detection.
x_link (pandas.DataFrame, optional) – The second DataFrame.
- Returns:
pandas.Series, pandas.DataFrame, numpy.ndarray – The result of comparing record pairs (the features). Can be a tuple with multiple pandas.Series, pandas.DataFrame, numpy.ndarray objects.
- class recordlinkage.compare.Geographic(left_on_lat, left_on_lng, right_on_lat, right_on_lng, method=None, offset=0.0, scale=1.0, origin=0.0, missing_value=0.0, label=None)
Compute the (partial) similarity between WGS84 coordinate values.
Compare the geometric (haversine) distance between two WGS- coordinates. The similarity algorithms are ‘step’, ‘linear’, ‘exp’, ‘gauss’ or ‘squared’. The similarity functions are the same as in
recordlinkage.comparing.Compare.numeric()- Parameters:
left_on_lat (tuple) – The name or position of the latitude in the left DataFrame.
left_on_lng (tuple) – The name or position of the longitude in the left DataFrame.
right_on_lat (tuple) – The name or position of the latitude in the right DataFrame.
right_on_lng (tuple) – The name or position of the longitude in the right DataFrame.
method (str) – The metric used. Options ‘step’, ‘linear’, ‘exp’, ‘gauss’ or ‘squared’. Default ‘linear’.
offset (float) – The offset. See Compare.numeric.
scale (float) – The scale of the numeric comparison method. See Compare.numeric. This argument is not available for the ‘step’ algorithm.
origin (float) – The shift of bias between the values. See Compare.numeric.
missing_value (numpy.dtype) – The value for a comparison with a missing value. Default 0.
- compute(pairs, x, x_link=None)
Compare the records of each record pair.
Calling this method starts the comparing of records.
- Parameters:
pairs (pandas.MultiIndex) – A pandas MultiIndex with the record pairs to compare. The indices in the MultiIndex are indices of the DataFrame(s) to link.
x (pandas.DataFrame) – The DataFrame to link. If x_link is given, the comparing is a linking problem. If x_link is not given, the problem is one of duplicate detection.
x_link (pandas.DataFrame, optional) – The second DataFrame.
- Returns:
pandas.Series, pandas.DataFrame, numpy.ndarray – The result of comparing record pairs (the features). Can be a tuple with multiple pandas.Series, pandas.DataFrame, numpy.ndarray objects.
- class recordlinkage.compare.Date(left_on, right_on, swap_month_day=0.5, swap_months='default', errors='coerce', missing_value=0.0, label=None)
Compute the (partial) similarity between date values.
- Parameters:
left_on (str or int) – The name or position of the column in the left DataFrame.
right_on (str or int) – The name or position of the column in the right DataFrame.
swap_month_day (float) – The value if the month and day are swapped. Default 0.5.
swap_months (list of tuples) – A list of tuples with common errors caused by the translating of months into numbers, i.e. October is month 10. The format of the tuples is (month_good, month_bad, value). Default : swap_months = [(6, 7, 0.5), (7, 6, 0.5), (9, 10, 0.5), (10, 9, 0.5)]
missing_value (numpy.dtype) – The value for a comparison with a missing value. Default 0.0.
- compute(pairs, x, x_link=None)
Compare the records of each record pair.
Calling this method starts the comparing of records.
- Parameters:
pairs (pandas.MultiIndex) – A pandas MultiIndex with the record pairs to compare. The indices in the MultiIndex are indices of the DataFrame(s) to link.
x (pandas.DataFrame) – The DataFrame to link. If x_link is given, the comparing is a linking problem. If x_link is not given, the problem is one of duplicate detection.
x_link (pandas.DataFrame, optional) – The second DataFrame.
- Returns:
pandas.Series, pandas.DataFrame, numpy.ndarray – The result of comparing record pairs (the features). Can be a tuple with multiple pandas.Series, pandas.DataFrame, numpy.ndarray objects.
- class recordlinkage.compare.Variable(left_on=None, right_on=None, missing_value=0.0, label=None)
Add a variable of the dataframe as feature.
- Parameters:
left_on (str or int) – The name or position of the column in the left DataFrame.
right_on (str or int) – The name or position of the column in the right DataFrame.
missing_value (numpy.dtype) – The value for a comparison with a missing value. Default 0.0.
- compute(pairs, x, x_link=None)
Compare the records of each record pair.
Calling this method starts the comparing of records.
- Parameters:
pairs (pandas.MultiIndex) – A pandas MultiIndex with the record pairs to compare. The indices in the MultiIndex are indices of the DataFrame(s) to link.
x (pandas.DataFrame) – The DataFrame to link. If x_link is given, the comparing is a linking problem. If x_link is not given, the problem is one of duplicate detection.
x_link (pandas.DataFrame, optional) – The second DataFrame.
- Returns:
pandas.Series, pandas.DataFrame, numpy.ndarray – The result of comparing record pairs (the features). Can be a tuple with multiple pandas.Series, pandas.DataFrame, numpy.ndarray objects.
- class recordlinkage.compare.VariableA(on=None, missing_value=0.0, label=None)
Add a variable of the left dataframe as feature.
- Parameters:
on (str or int) – The name or position of the column in the left DataFrame.
normalise (bool) – Normalise the outcome. This is needed for good result in many classification models. Default True.
missing_value (numpy.dtype) – The value for a comparison with a missing value. Default 0.0.
- compute(pairs, x, x_link=None)
Compare the records of each record pair.
Calling this method starts the comparing of records.
- Parameters:
pairs (pandas.MultiIndex) – A pandas MultiIndex with the record pairs to compare. The indices in the MultiIndex are indices of the DataFrame(s) to link.
x (pandas.DataFrame) – The DataFrame to link. If x_link is given, the comparing is a linking problem. If x_link is not given, the problem is one of duplicate detection.
x_link (pandas.DataFrame, optional) – The second DataFrame.
- Returns:
pandas.Series, pandas.DataFrame, numpy.ndarray – The result of comparing record pairs (the features). Can be a tuple with multiple pandas.Series, pandas.DataFrame, numpy.ndarray objects.
- class recordlinkage.compare.VariableB(on=None, missing_value=0.0, label=None)
Add a variable of the right dataframe as feature.
- Parameters:
on (str or int) – The name or position of the column in the right DataFrame.
normalise (bool) – Normalise the outcome. This is needed for good result in many classification models. Default True.
missing_value (numpy.dtype) – The value for a comparison with a missing value. Default 0.0.
- compute(pairs, x, x_link=None)
Compare the records of each record pair.
Calling this method starts the comparing of records.
- Parameters:
pairs (pandas.MultiIndex) – A pandas MultiIndex with the record pairs to compare. The indices in the MultiIndex are indices of the DataFrame(s) to link.
x (pandas.DataFrame) – The DataFrame to link. If x_link is given, the comparing is a linking problem. If x_link is not given, the problem is one of duplicate detection.
x_link (pandas.DataFrame, optional) – The second DataFrame.
- Returns:
pandas.Series, pandas.DataFrame, numpy.ndarray – The result of comparing record pairs (the features). Can be a tuple with multiple pandas.Series, pandas.DataFrame, numpy.ndarray objects.
- class recordlinkage.compare.Frequency(left_on=None, right_on=None, normalise=True, missing_value=0.0, label=None)
Compute the (relative) frequency of each variable.
- Parameters:
left_on (str or int) – The name or position of the column in the left DataFrame.
right_on (str or int) – The name or position of the column in the right DataFrame.
normalise (bool) – Normalise the outcome. This is needed for good result in many classification models. Default True.
missing_value (numpy.dtype) – The value for a comparison with a missing value. Default 0.0.
- compute(pairs, x, x_link=None)
Compare the records of each record pair.
Calling this method starts the comparing of records.
- Parameters:
pairs (pandas.MultiIndex) – A pandas MultiIndex with the record pairs to compare. The indices in the MultiIndex are indices of the DataFrame(s) to link.
x (pandas.DataFrame) – The DataFrame to link. If x_link is given, the comparing is a linking problem. If x_link is not given, the problem is one of duplicate detection.
x_link (pandas.DataFrame, optional) – The second DataFrame.
- Returns:
pandas.Series, pandas.DataFrame, numpy.ndarray – The result of comparing record pairs (the features). Can be a tuple with multiple pandas.Series, pandas.DataFrame, numpy.ndarray objects.
- class recordlinkage.compare.FrequencyA(on=None, normalise=True, missing_value=0.0, label=None)
Compute the frequency of a variable in the left dataframe.
- Parameters:
on (str or int) – The name or position of the column in the left DataFrame.
normalise (bool) – Normalise the outcome. This is needed for good result in many classification models. Default True.
missing_value (numpy.dtype) – The value for a comparison with a missing value. Default 0.0.
- compute(pairs, x, x_link=None)
Compare the records of each record pair.
Calling this method starts the comparing of records.
- Parameters:
pairs (pandas.MultiIndex) – A pandas MultiIndex with the record pairs to compare. The indices in the MultiIndex are indices of the DataFrame(s) to link.
x (pandas.DataFrame) – The DataFrame to link. If x_link is given, the comparing is a linking problem. If x_link is not given, the problem is one of duplicate detection.
x_link (pandas.DataFrame, optional) – The second DataFrame.
- Returns:
pandas.Series, pandas.DataFrame, numpy.ndarray – The result of comparing record pairs (the features). Can be a tuple with multiple pandas.Series, pandas.DataFrame, numpy.ndarray objects.
- class recordlinkage.compare.FrequencyB(on=None, normalise=True, missing_value=0.0, label=None)
Compute the frequency of a variable in the right dataframe.
- Parameters:
on (str or int) – The name or position of the column in the right DataFrame.
normalise (bool) – Normalise the outcome. This is needed for good result in many classification models. Default True.
missing_value (numpy.dtype) – The value for a comparison with a missing value. Default 0.0.
- compute(pairs, x, x_link=None)
Compare the records of each record pair.
Calling this method starts the comparing of records.
- Parameters:
pairs (pandas.MultiIndex) – A pandas MultiIndex with the record pairs to compare. The indices in the MultiIndex are indices of the DataFrame(s) to link.
x (pandas.DataFrame) – The DataFrame to link. If x_link is given, the comparing is a linking problem. If x_link is not given, the problem is one of duplicate detection.
x_link (pandas.DataFrame, optional) – The second DataFrame.
- Returns:
pandas.Series, pandas.DataFrame, numpy.ndarray – The result of comparing record pairs (the features). Can be a tuple with multiple pandas.Series, pandas.DataFrame, numpy.ndarray objects.
User-defined algorithms
A user-defined algorithm can be defined based on
recordlinkage.base.BaseCompareFeature. The recordlinkage.base.BaseCompareFeature class is an abstract base
class that is used for compare algorithms. The classes
are inherited from this abstract base class. You can use BaseCompareFeature to
create a user-defined/custom algorithm. Overwrite the abstract method
recordlinkage.base.BaseCompareFeature._compute_vectorized() with the
compare algorithm. A short example is given here:
from recordlinkage.base import BaseCompareFeature
class CustomFeature(BaseCompareFeature):
def _compute_vectorized(s1, s2):
# algorithm that compares s1 and s2
# return a pandas.Series
return ...
feat = CustomFeature()
feat.compute(pairs, dfA, dfB)
A full description of the recordlinkage.base.BaseCompareFeature
class:
- class recordlinkage.base.BaseCompareFeature(labels_left, labels_right, args=(), kwargs={}, label=None)
Base abstract class for compare feature engineering.
- Parameters:
labels_left (list, str, int) – The labels to use for comparing record pairs in the left dataframe.
labels_right (list, str, int) – The labels to use for comparing record pairs in the right dataframe (linking) or left dataframe (duplicate detection).
args (tuple) – Additional arguments to pass to the _compare_vectorized method.
kwargs (tuple) – Keyword additional arguments to pass to the _compare_vectorized method.
label (list, str, int) – The identifying label(s) for the returned values.
- compute(pairs, x, x_link=None)
Compare the records of each record pair.
Calling this method starts the comparing of records.
- Parameters:
pairs (pandas.MultiIndex) – A pandas MultiIndex with the record pairs to compare. The indices in the MultiIndex are indices of the DataFrame(s) to link.
x (pandas.DataFrame) – The DataFrame to link. If x_link is given, the comparing is a linking problem. If x_link is not given, the problem is one of duplicate detection.
x_link (pandas.DataFrame, optional) – The second DataFrame.
- Returns:
pandas.Series, pandas.DataFrame, numpy.ndarray – The result of comparing record pairs (the features). Can be a tuple with multiple pandas.Series, pandas.DataFrame, numpy.ndarray objects.
- _compute(left_on, right_on)
Compare the data on the left and right.
BaseCompareFeature._compute()andBaseCompareFeature.compute()differ on the accepted arguments. _compute accepts indexed data while compute accepts the record pairs and the DataFrame’s.- Parameters:
left_on ((tuple of) pandas.Series) – Data to compare with right_on
right_on ((tuple of) pandas.Series) – Data to compare with left_on
- Returns:
pandas.Series, pandas.DataFrame, numpy.ndarray – The result of comparing record pairs (the features). Can be a tuple with multiple pandas.Series, pandas.DataFrame, numpy.ndarray objects.
- _compute_vectorized(*args)
Compare attributes (vectorized)
- Parameters:
*args (pandas.Series) – pandas.Series’ as arguments.
- Returns:
pandas.Series, pandas.DataFrame, numpy.ndarray – The result of comparing record pairs (the features). Can be a tuple with multiple pandas.Series, pandas.DataFrame, numpy.ndarray objects.
Warning
Do not change the order of the pairs in the MultiIndex.
Examples
Example: High level usage
import recordlinkage as rl
comparer = rl.Compare()
comparer.string('name_a', 'name_b', method='jarowinkler', threshold=0.85, label='name')
comparer.exact('sex', 'gender', label='gender')
comparer.date('dob', 'date_of_birth', label='date')
comparer.string('str_name', 'streetname', method='damerau_levenshtein', threshold=0.7, label='streetname')
comparer.exact('place', 'placename', label='placename')
comparer.numeric('income', 'income', method='gauss', offset=3, scale=3, missing_value=0.5, label='income')
comparer.compute(pairs, dfA, dfB)
Example: Low level usage
import recordlinkage as rl
from recordlinkage.compare import Exact, String, Numeric, Date
comparer = rl.Compare([
String('name_a', 'name_b', method='jarowinkler', threshold=0.85, label='name'),
Exact('sex', 'gender', label='gender'),
Date('dob', 'date_of_birth', label='date'),
String('str_name', 'streetname', method='damerau_levenshtein', threshold=0.7, label='streetname'),
Exact('place', 'placename', label='placename'),
Numeric('income', 'income', method='gauss', offset=3, scale=3, missing_value=0.5, label='income'),
])
comparer.compute(pairs, dfA, dfB)
The following examples give a feeling on the extensibility of the toolkit.
Example: User-defined algorithm 1
The following code defines a custom algorithm to compare zipcodes. The algorithm returns 1.0 for record pairs that agree on the zipcode and returns 0.0 for records that disagree on the zipcode. If the zipcodes disagree but the first two numbers are identical, then the algorithm returns 0.5.
import recordlinkage as rl
from recordlinkage.base import BaseCompareFeature
class CompareZipCodes(BaseCompareFeature):
def _compute_vectorized(self, s1, s2):
"""Compare zipcodes.
If the zipcodes in both records are identical, the similarity
is 1. If the first two values agree and the last two don't, then
the similarity is 0.5. Otherwise, the similarity is 0.
"""
# check if the zipcode are identical (return 1 or 0)
sim = (s1 == s2).astype(float)
# check the first 2 numbers of the distinct comparisons
sim[(sim == 0) & (s1.str[0:2] == s2.str[0:2])] = 0.5
return sim
comparer = rl.Compare()
comparer.extact('given_name', 'given_name', 'y_name')
comparer.string('surname', 'surname', 'y_surname')
comparer.add(CompareZipCodes('postcode', 'postcode', label='y_postcode'))
comparer.compute(pairs, dfA, dfB)
0.0 71229
0.5 3166
1.0 2854
Name: sim_postcode, dtype: int64
Note
See recordlinkage.base.BaseCompareFeature for more
details on how to subclass.
Example: User-defined algorithm 2
As you can see, one can pass the labels of the columns as arguments. The
first argument is a column label, or a list of column labels, found in
the first DataFrame (postcode in this example). The second argument
is a column label, or a list of column labels, found in the second
DataFrame (also postcode in this example). The
recordlinkage.Compare class selects the columns with the given
labels before passing them to the custom algorithm/function. The
compare method in the recordlinkage.Compare class passes
additional (keyword) arguments to the custom function.
Warning: Do not change the order of the pairs in the MultiIndex.
import recordlinkage as rl
from recordlinkage.base import BaseCompareFeature
class CompareZipCodes(BaseCompareFeature):
def __init__(self, left_on, right_on, partial_sim_value, *args, **kwargs):
super(CompareZipCodes, self).__init__(left_on, right_on, *args, **kwargs)
self.partial_sim_value = partial_sim_value
def _compute_vectorized(self, s1, s2):
"""Compare zipcodes.
If the zipcodes in both records are identical, the similarity
is 0. If the first two values agree and the last two don't, then
the similarity is 0.5. Otherwise, the similarity is 0.
"""
# check if the zipcode are identical (return 1 or 0)
sim = (s1 == s2).astype(float)
# check the first 2 numbers of the distinct comparisons
sim[(sim == 0) & (s1.str[0:2] == s2.str[0:2])] = self.partial_sim_value
return sim
comparer = rl.Compare()
comparer.extact('given_name', 'given_name', 'y_name')
comparer.string('surname', 'surname', 'y_surname')
comparer.add(CompareZipCodes('postcode', 'postcode',
'partial_sim_value'=0.5, label='y_postcode'))
comparer.compute(pairs, dfA, dfB)
Example: User-defined algorithm 3
The Python Record Linkage Toolkit supports the comparison of more than two columns. This is especially useful in situations with multi-dimensional data (for example geographical coordinates) and situations where fields can be swapped.
The FEBRL4 dataset has two columns filled with address information
(address_1 and address_2). In a naive approach, one compares
address_1 of file A with address_1 of file B and address_2
of file A with address_2 of file B. If the values for address_1
and address_2 are swapped during the record generating process, the
naive approach considers the addresses to be distinct. In a more
advanced approach, address_1 of file A is compared with
address_1 and address_2 of file B. Variable address_2 of
file A is compared with address_1 and address_2 of file B. This
is done with the single function given below.
import recordlinkage as rl
from recordlinkage.base import BaseCompareFeature
class CompareAddress(BaseCompareFeature):
def _compute_vectorized(self, s1_1, s1_2, s2_1, s2_2):
"""Compare addresses.
Compare addresses. Compare address_1 of file A with
address_1 and address_2 of file B. The same for address_2
of dataset 1.
"""
return ((s1_1 == s2_1) | (s1_2 == s2_2) | (s1_1 == s2_2) | (s1_2 == s2_1)).astype(float)
comparer = rl.Compare()
# naive
comparer.add(CompareAddress('address_1', 'address_1', label='sim_address_1'))
comparer.add(CompareAddress('address_2', 'address_2', label='sim_address_2'))
# better
comparer.add(CompareAddress(('address_1', 'address_2'),
('address_1', 'address_2'),
label='sim_address'
)
features = comparer.compute(pairs, dfA, dfB)
features.mean()
The mean of the cross-over comparison is higher.
sim_address_1 0.02488
sim_address_2 0.02025
sim_address 0.03566
dtype: float64